Cassava Leaf Disease - Exploratory Data Analysis
import numpy as np
import pandas as pdimport os
BASE_DIR = "../input"
for dirname, _, filenames in os.walk(BASE_DIR):
for filename in filenames:
print(os.path.join(dirname, filename))dirname,_,filename('../input\\cassava-leaf-disease-classification\\train_tfrecords',
[],
'ld_train15-1327.tfrec')BASE_DIR = "../input/cassava-leaf-disease-classification/"import json
label_map = []
with open(BASE_DIR + "label_num_to_disease_map.json", 'r') as f:
for line in f:
label_map.append(json.loads(line))
label_map = pd.DataFrame(label_map)
label_map| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | Cassava Bacterial Blight (CBB) | Cassava Brown Streak Disease (CBSD) | Cassava Green Mottle (CGM) | Cassava Mosaic Disease (CMD) | Healthy |
df_train = pd.read_csv(BASE_DIR + "train.csv")
print(len(df_train))
df_train.head()21397| image_id | label | |
|---|---|---|
| 0 | 1000015157.jpg | 0 |
| 1 | 1000201771.jpg | 3 |
| 2 | 100042118.jpg | 1 |
| 3 | 1000723321.jpg | 1 |
| 4 | 1000812911.jpg | 3 |
df_train["label"][1]3type(label_map.iloc[0,3])str# 使用 for 循环实现增加一列 class_name
# for i in range(len(df_train)):
# df_train["class_name"][i] = label_map.iloc[0,df_train["label"][i]]
# df_train.head()df_train["label"].apply(lambda x: label_map.iloc[0,x])0 Cassava Bacterial Blight (CBB)
1 Cassava Mosaic Disease (CMD)
2 Cassava Brown Streak Disease (CBSD)
3 Cassava Brown Streak Disease (CBSD)
4 Cassava Mosaic Disease (CMD)
...
21392 Cassava Mosaic Disease (CMD)
21393 Cassava Mosaic Disease (CMD)
21394 Cassava Brown Streak Disease (CBSD)
21395 Healthy
21396 Healthy
Name: label, Length: 21397, dtype: objectdf_train["class_name"] = df_train["label"].apply(lambda x: label_map.iloc[0,x])
df_train.head()| image_id | label | class_name | |
|---|---|---|---|
| 0 | 1000015157.jpg | 0 | Cassava Bacterial Blight (CBB) |
| 1 | 1000201771.jpg | 3 | Cassava Mosaic Disease (CMD) |
| 2 | 100042118.jpg | 1 | Cassava Brown Streak Disease (CBSD) |
| 3 | 1000723321.jpg | 1 | Cassava Brown Streak Disease (CBSD) |
| 4 | 1000812911.jpg | 3 | Cassava Mosaic Disease (CMD) |
df_train["class_name_"] = df_train["label"].map(lambda x: label_map.iloc[0,x])
df_train| image_id | label | class_name | class_name_ | |
|---|---|---|---|---|
| 0 | 1000015157.jpg | 0 | Cassava Bacterial Blight (CBB) | Cassava Bacterial Blight (CBB) |
| 1 | 1000201771.jpg | 3 | Cassava Mosaic Disease (CMD) | Cassava Mosaic Disease (CMD) |
| 2 | 100042118.jpg | 1 | Cassava Brown Streak Disease (CBSD) | Cassava Brown Streak Disease (CBSD) |
| 3 | 1000723321.jpg | 1 | Cassava Brown Streak Disease (CBSD) | Cassava Brown Streak Disease (CBSD) |
| 4 | 1000812911.jpg | 3 | Cassava Mosaic Disease (CMD) | Cassava Mosaic Disease (CMD) |
| ... | ... | ... | ... | ... |
| 21392 | 999068805.jpg | 3 | Cassava Mosaic Disease (CMD) | Cassava Mosaic Disease (CMD) |
| 21393 | 999329392.jpg | 3 | Cassava Mosaic Disease (CMD) | Cassava Mosaic Disease (CMD) |
| 21394 | 999474432.jpg | 1 | Cassava Brown Streak Disease (CBSD) | Cassava Brown Streak Disease (CBSD) |
| 21395 | 999616605.jpg | 4 | Healthy | Healthy |
| 21396 | 999998473.jpg | 4 | Healthy | Healthy |
21397 rows × 4 columns
df_train = df_train.drop(['class_name_'], axis = 1)
df_train.head()| image_id | label | class_name | |
|---|---|---|---|
| 0 | 1000015157.jpg | 0 | Cassava Bacterial Blight (CBB) |
| 1 | 1000201771.jpg | 3 | Cassava Mosaic Disease (CMD) |
| 2 | 100042118.jpg | 1 | Cassava Brown Streak Disease (CBSD) |
| 3 | 1000723321.jpg | 1 | Cassava Brown Streak Disease (CBSD) |
| 4 | 1000812911.jpg | 3 | Cassava Mosaic Disease (CMD) |
import cv2
img_shape = {}
for image_name in os.listdir(BASE_DIR + "train_images")[:300]:
image = cv2.imread(BASE_DIR + "train_images/" + image_name)
img_shape[image.shape] = img_shape.get(image.shape, 0) + 1
print(img_shape){(600, 800, 3): 300}print(image_name, type(image))1052903541.jpg <class 'numpy.ndarray'># 使用 cv2 查看图片
# cv2.imshow("figure",image)
# cv2.waitKey(0)字典 get 函数
描述
Python 字典 get() 函数返回指定键的值,如果值不在字典中返回默认值。
语法
get()方法语法:
dict.get(key, default=None)
参数
key – 字典中要查找的键。
default – 如果指定键的值不存在时,返回该默认值值。
返回值 - 返回指定键的值,如果值不在字典中返回默认值 None。
dict_ = {'Name': "Terence", 'Age': 19}
print("Age 的值为", dict_.get('Age'))
print("Sex 的值为", dict_.get('Sex'))
print("Sex 的值为", dict_.get('Sex',0))Age 的值为 19
Sex 的值为 None
Sex 的值为 0seaborn 绘制类别分布比例
seaborn.countplot
Bar graphs are useful for displaying relationships between categorical data and at least one numerical variable. seaborn.countplot is a barplot where the dependent variable is the number of instances of each instance of the independent variable.
import matplotlib.pyplot as plt
import seaborn as sn
fig, axes = plt.subplots(figsize=(8,4))
sn.countplot(y = "class_name", data = df_train);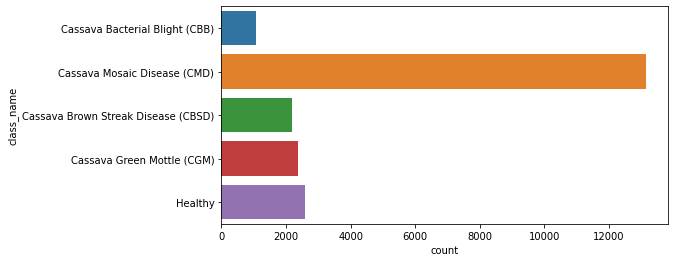
df_train.head()| image_id | label | class_name | |
|---|---|---|---|
| 0 | 1000015157.jpg | 0 | Cassava Bacterial Blight (CBB) |
| 1 | 1000201771.jpg | 3 | Cassava Mosaic Disease (CMD) |
| 2 | 100042118.jpg | 1 | Cassava Brown Streak Disease (CBSD) |
| 3 | 1000723321.jpg | 1 | Cassava Brown Streak Disease (CBSD) |
| 4 | 1000812911.jpg | 3 | Cassava Mosaic Disease (CMD) |
type(df_train["image_id"]),type(df_train["image_id"].values)(pandas.core.series.Series, numpy.ndarray)tmp_df = df_train.sample(9)
image_ids = tmp_df["image_id"].values
labels = tmp_df["class_name"].valuesfor index, (image_id, label) in enumerate(zip(image_ids, labels)):
print(index, image_id, label)0 3594707809.jpg Cassava Mosaic Disease (CMD)
1 498735095.jpg Cassava Mosaic Disease (CMD)
2 597389720.jpg Cassava Green Mottle (CGM)
3 232417860.jpg Cassava Mosaic Disease (CMD)
4 3969928849.jpg Healthy
5 1111878443.jpg Cassava Mosaic Disease (CMD)
6 2263743432.jpg Cassava Bacterial Blight (CBB)
7 965919968.jpg Cassava Bacterial Blight (CBB)
8 2418850424.jpg Cassava Mosaic Disease (CMD)# def visualize_batch_(image_ids, labels):
# plt.figure(figsize=(16, 12))
# for ind, (image_id, label) in enumerate(zip(image_ids, labels)):
# plt.subplot(3, 3, ind + 1)
# image = cv2.imread(os.path.join(BASE_DIR, "train_images", image_id))
# image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# plt.imshow(image)
# plt.title(f"Class: {label}", fontsize=12)
# plt.axis("off")
# plt.show()# visualize_batch_(image_ids, labels)ax:matplotlib.axes._subplots.AxesSubplot,的基本操作
ax.set_xticks([]), ax_set_yticks([]):关闭坐标刻度ax.axis('off'):关闭坐标轴ax.set_title():设置标题
def visulaize_batch(image_ids, labels):
fig, axes = plt.subplots(3, 3, figsize=(16,12))
for index, (image_id, label) in enumerate(zip(image_ids, labels)):
# 下面两个 image 类型一致,都是 <class 'numpy.ndarray'>
image = cv2.imread(BASE_DIR + "train_images/" + image_id)
# print(type(image))
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# print(type(image))
# 这里需要对 axes 进行索引,确定其具体位置,而且必须使用二维索引如:axes[2,1],而不能是 axes[6]
axes[index//3,index%3].imshow(image)
axes[index//3,index%3].set_title(f"Class: {label}", fontsize = 12)
axes[index//3,index%3].axis("off") #去除坐标轴信息
plt.show()visulaize_batch(image_ids, labels)
0 - CBB - Cassava Bacterial Blight
tmp_df = df_train[df_train["label"] == 0]
print(f"Total train image for class 0(Cassava Bacterial Blight): {tmp_df.shape[0]}")
tmp_df = tmp_df.sample(9)
image_ids = tmp_df["image_id"].values
labels = tmp_df["label"].values
visulaize_batch(image_ids, labels)Total train image for class 0(Cassava Bacterial Blight): 1087
1 - CBSD - Cassava Brown Streak Disease
tmp_df = df_train[df_train["label"] == 1]
print(f"Total train image for class 1(Cassava Brown Streak Disease): {tmp_df.shape[0]}")
tmp_df = tmp_df.sample(9)
image_ids = tmp_df["image_id"].values
labels = tmp_df["label"].values
visulaize_batch(image_ids, labels)Total train image for class 1(Cassava Brown Streak Disease): 2189
2 - CGM - Cassava Green Mottle
tmp_df = df_train[df_train["label"] == 2]
print(f"Total train image for class 2(Cassava Green Mottle): {tmp_df.shape[0]}")
tmp_df = tmp_df.sample(9)
image_ids = tmp_df["image_id"].values
labels = tmp_df["label"].values
visulaize_batch(image_ids, labels)Total train image for class 2(Cassava Green Mottle): 2386
3 - CMD - Cassava Mosaic Disease
tmp_df = df_train[df_train["label"] == 3]
print(f"Total train image for class 3(Cassava Mosaic Disease): {tmp_df.shape[0]}")
tmp_df = tmp_df.sample(9)
image_ids = tmp_df["image_id"].values
labels = tmp_df["label"].values
visulaize_batch(image_ids, labels)Total train image for class 3(Cassava Mosaic Disease): 13158
4 - Healthy
tmp_df = df_train[df_train["label"] == 4]
print(f"Total train image for class 4(Healthy): {tmp_df.shape[0]}")
tmp_df = tmp_df.sample(9)
image_ids = tmp_df["image_id"].values
labels = tmp_df["label"].values
visulaize_batch(image_ids, labels)Total train image for class 4(Healthy): 2577
albumentations数据增强
albumentations 是一个给予 OpenCV的快速训练数据增强库,拥有非常简单且强大的可以用于多种任务(分割、检测)的接口,易于定制且添加其他框架非常方便。
它可以对数据集进行逐像素的转换,如模糊、下采样、高斯造点、高斯模糊、动态模糊、RGB 转换、随机雾化等;也可以进行空间转换(同时也会对目标进行转换),如裁剪、翻转、随机裁剪等。
github 及其示例地址如下:
- GitHub: https://github.com/albumentations-team/albumentations
- 示例:https://github.com/albumentations-team/albumentations_examples
def plot_augmentation(image_id, transform):
fig, axes = plt.subplots(1, 3, figsize=(16,4))
image = cv2.imread(BASE_DIR + "train_images/" + image_id)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
axes[0].imshow(image)
axes[0].axis("off")
x = transform(image = image)["image"]
axes[1].imshow(x)
axes[1].axis("off")
x = transform(image = image)["image"]
axes[2].imshow(x)
axes[2].axis("off")
plt.show()import albumentations as A
transform_shift_scale_rotate = A.ShiftScaleRotate(
p = 1.0,
shift_limit = (-0.3, 0.3),
scale_limit = (-0.1, 0.1),
rotate_limit = (-180, 180),
interpolation = 0,
border_mode = 4,
)
df_train['image_id'][0],type(df_train['image_id'][0])('1000015157.jpg', str)plot_augmentation(df_train['image_id'][2], transform_shift_scale_rotate)
transform_coarse_dropout = A.CoarseDropout(
p=1.0,
max_holes=100,
max_height=50,
max_width=50,
min_holes=30,
min_height=20,
min_width=20,
)
plot_augmentation(df_train['image_id'][2], transform_coarse_dropout)
transform = A.Compose(
transforms=[
transform_shift_scale_rotate,
transform_coarse_dropout,
],
p=1.0,
)
plot_augmentation(df_train['image_id'][2], transform)
ResNet50 看聚类
df_train.head()| image_id | label | class_name | |
|---|---|---|---|
| 0 | 1000015157.jpg | 0 | Cassava Bacterial Blight (CBB) |
| 1 | 1000201771.jpg | 3 | Cassava Mosaic Disease (CMD) |
| 2 | 100042118.jpg | 1 | Cassava Brown Streak Disease (CBSD) |
| 3 | 1000723321.jpg | 1 | Cassava Brown Streak Disease (CBSD) |
| 4 | 1000812911.jpg | 3 | Cassava Mosaic Disease (CMD) |
from keras.preprocessing.image import load_img
from keras.applications.resnet50 import preprocess_input
def extract_features(image_id, model):
file = BASE_DIR + "train_images/" + image_id
# load the image as a 224*224 array
img = load_img(file, target_size=(224, 224))
# turn to numpy.array
img = np.array(img)
# reshape the data for the model shape = (num_of_samples, dim1, dim2, channels)
reshaped_img = img.reshape(1, 224, 224, 3)
#
imgx = preprocess_input(reshaped_img)
features = model.predict(imgx, use_multiprocessing=True)
return featuresfrom keras.applications.resnet50 import ResNet50
from keras.models import Model
from tqdm import tqdm
model = ResNet50()
model = Model(inputs = model.inputs, outputs = model.layers[-2].output)
healthy = df_train[df_train['label'] == 4]
healthy['features'] = healthy['image_id'].apply(lambda x: extract_features(x, model))<ipython-input-51-f5196e3b723b>:9: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
healthy['features'] = healthy['image_id'].apply(lambda x: extract_features(x, model))healthy| image_id | label | class_name | features | |
|---|---|---|---|---|
| 8 | 1001723730.jpg | 4 | Healthy | [[0.35207286, 0.0, 1.1103351, 0.009539512, 0.0... |
| 17 | 1003442061.jpg | 4 | Healthy | [[3.8534477, 0.0, 0.06910457, 0.23157704, 0.0,... |
| 30 | 100560400.jpg | 4 | Healthy | [[0.23260176, 0.0028819551, 2.424273, 0.0, 0.0... |
| 47 | 1009126931.jpg | 4 | Healthy | [[3.6958485, 0.1106378, 0.057800114, 0.1620144... |
| 62 | 1010806468.jpg | 4 | Healthy | [[3.0952048, 0.0, 0.0, 0.017503712, 0.0, 0.020... |
| ... | ... | ... | ... | ... |
| 21367 | 993984792.jpg | 4 | Healthy | [[0.8686834, 0.0, 0.04545398, 0.55075073, 0.0,... |
| 21372 | 995075067.jpg | 4 | Healthy | [[0.62431216, 0.0, 0.4463164, 0.11152529, 0.0,... |
| 21373 | 995123333.jpg | 4 | Healthy | [[0.0, 0.0, 0.41213, 0.23793063, 0.0, 0.0, 0.1... |
| 21395 | 999616605.jpg | 4 | Healthy | [[0.26363245, 0.0, 0.008111278, 0.348491, 0.0,... |
| 21396 | 999998473.jpg | 4 | Healthy | [[1.8681935, 0.0, 2.4172454, 0.21481319, 0.0, ... |
2577 rows × 4 columns
from sklearn.cluster import KMeans
features = np.array(healthy['features'].values.tolist()).reshape(-1,2048)
image_ids = np.array(healthy['image_id'].values.tolist())
# Clustering
kmeans = KMeans(n_clusters=5,n_jobs=-1, random_state=22)
kmeans.fit(features)C:\Users\Administrator\Desktop\Competition_Data_2020\zy\Code\venv\lib\site-packages\sklearn\cluster\_kmeans.py:938: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 0.25.
warnings.warn("'n_jobs' was deprecated in version 0.23 and will be"
KMeans(n_clusters=5, n_jobs=-1, random_state=22


